Tutorial for cropPAL2020
The cropPAL2020 home page
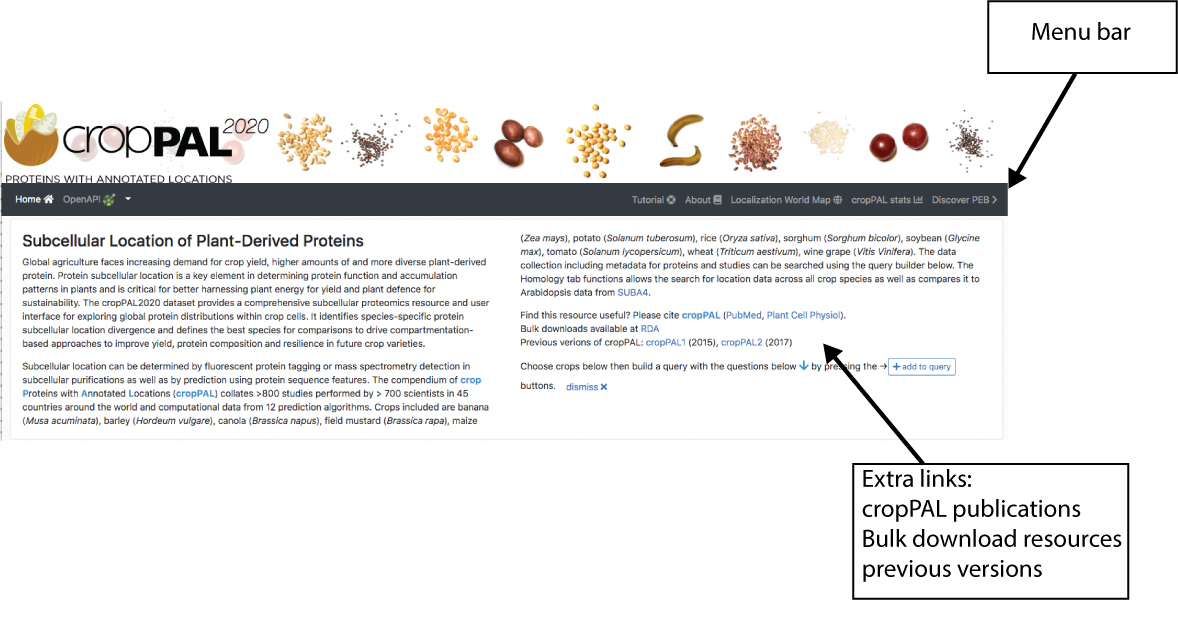
The home page view of cropPAL (http://crop-pal.org) provides the user with a introductory summary, links in the menu bar as well as extra links to bulk download, previous releases and most recent publication for citing cropPAL.
The search window below the summary is the starting point to the quick search of the query builder.
Quick search:
- Choose your target species
- Enter keywords to text-search for gene description annotations, titles or abstracts of studies or protein ids for a specific ID-search
Query Builder
The bottom part of the page contains the query builder tabs for creating more complex queries regarding subcellular locations, protein properties, homology or BLAST sequences against the cropPAL collection.

- Choose your target species (one or more)
- Choose your tab (Subcellular Location, Protein Properties, Homology, Publications or BLAST)
- Fill out a sentence and add it to the query with the button.
The Menu links
The links in the upper menu contain overview information about what is in cropPAL.

About
The about pages contains details about the built of different aspects of cropPAL including the genome annotations, methodology of data generation and other useful information.
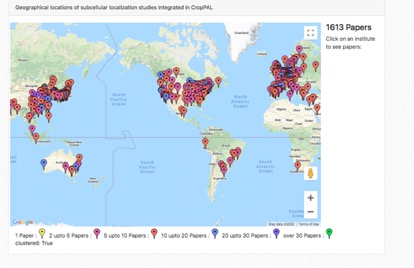
World map
The world map view described were all the data in cropPAL is derived from. Each flag symbolizes the affiliation from a distinct paper. The Flags are colour- coded to illustrate how many time the institution has contributed to the cropPAL data set.
The data display also includes protein-protein interaction studies performed in Arabidopsis that has been utilized in cropPAL to infer possible protein-protein interactions in crop species.
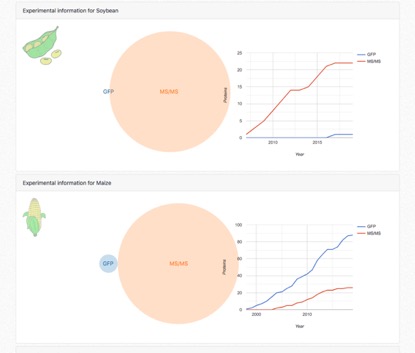
cropPAL stats
Illustrates the integrated experimental studies for each species over the years of publications. This indicates trends of subcellular research for individual species. The data is split into FP and MSMS derived experimental data sets.
Discover PEB
The link to the ARC Centre of Excellence in Plant Energy Biology. Explore the research, the people and the other data resource we can provide.
Search options
The Quick Search option
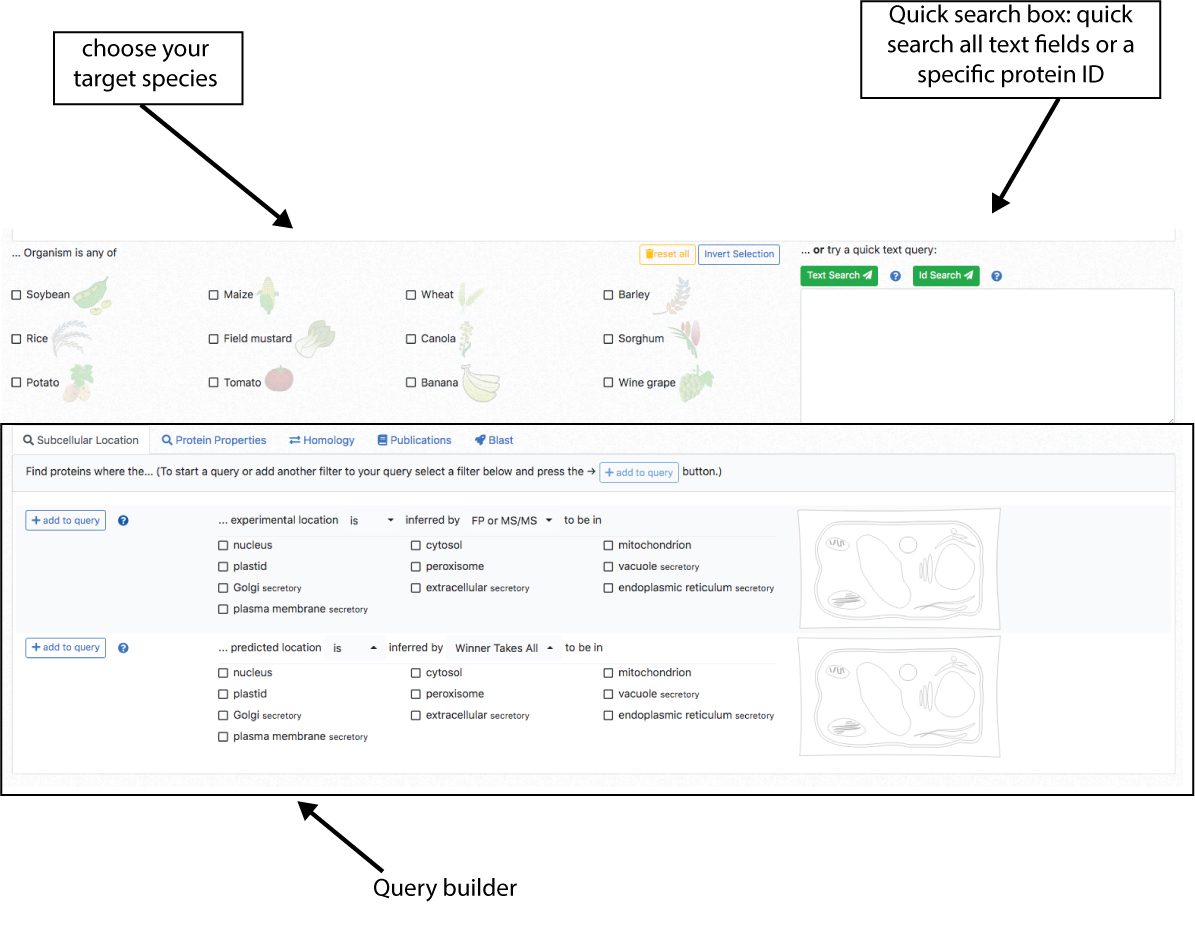
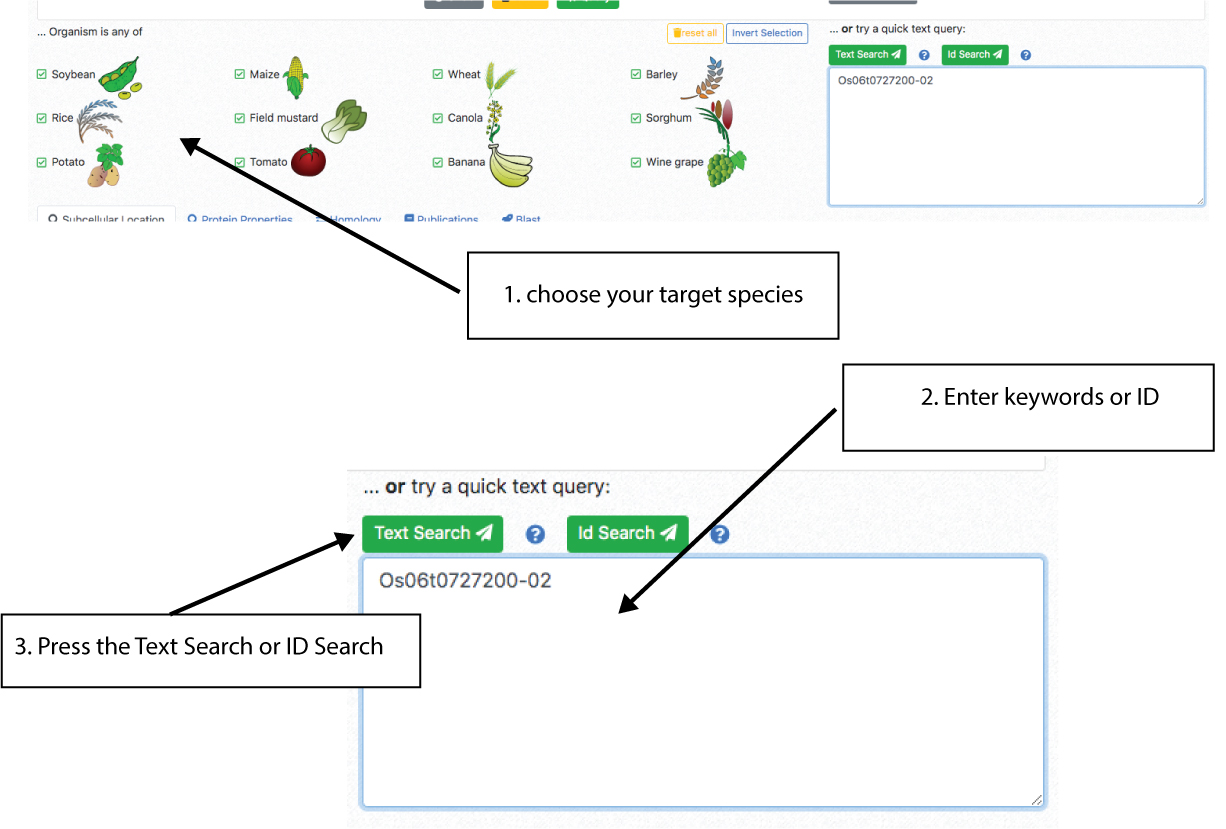
The first step for any search is choosing the crop species that will be queried. The crop species are shown on the left.
It is possible to choose from one to all crops. Then, in order to do a quick search us the Quick search window on the right.
This search will allow you to search for keywords in the gene description and title or abstracts of experimental studies. Enter you keyword (e.g. peroxidase) and press the text search button.
Alternatively, you can search for protein identifiers (e.g. Os06t0727200-02) to retrieve cropPAL data for the matching entries.
Note: The ID search is very specific and only finds exact protein IDs
matching the Ensembl ID system and version.
To search gene IDs or other IDs use the text search or the protein property tab.
The Query Builder
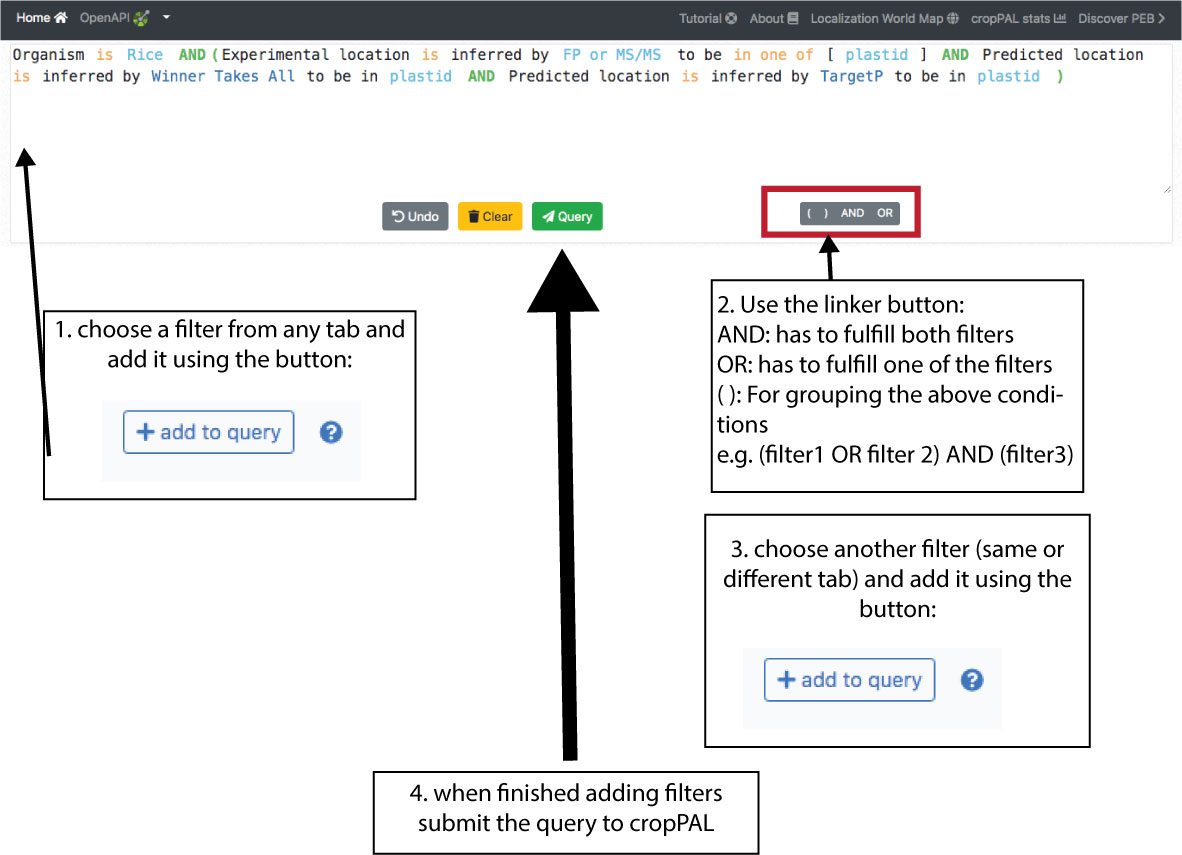
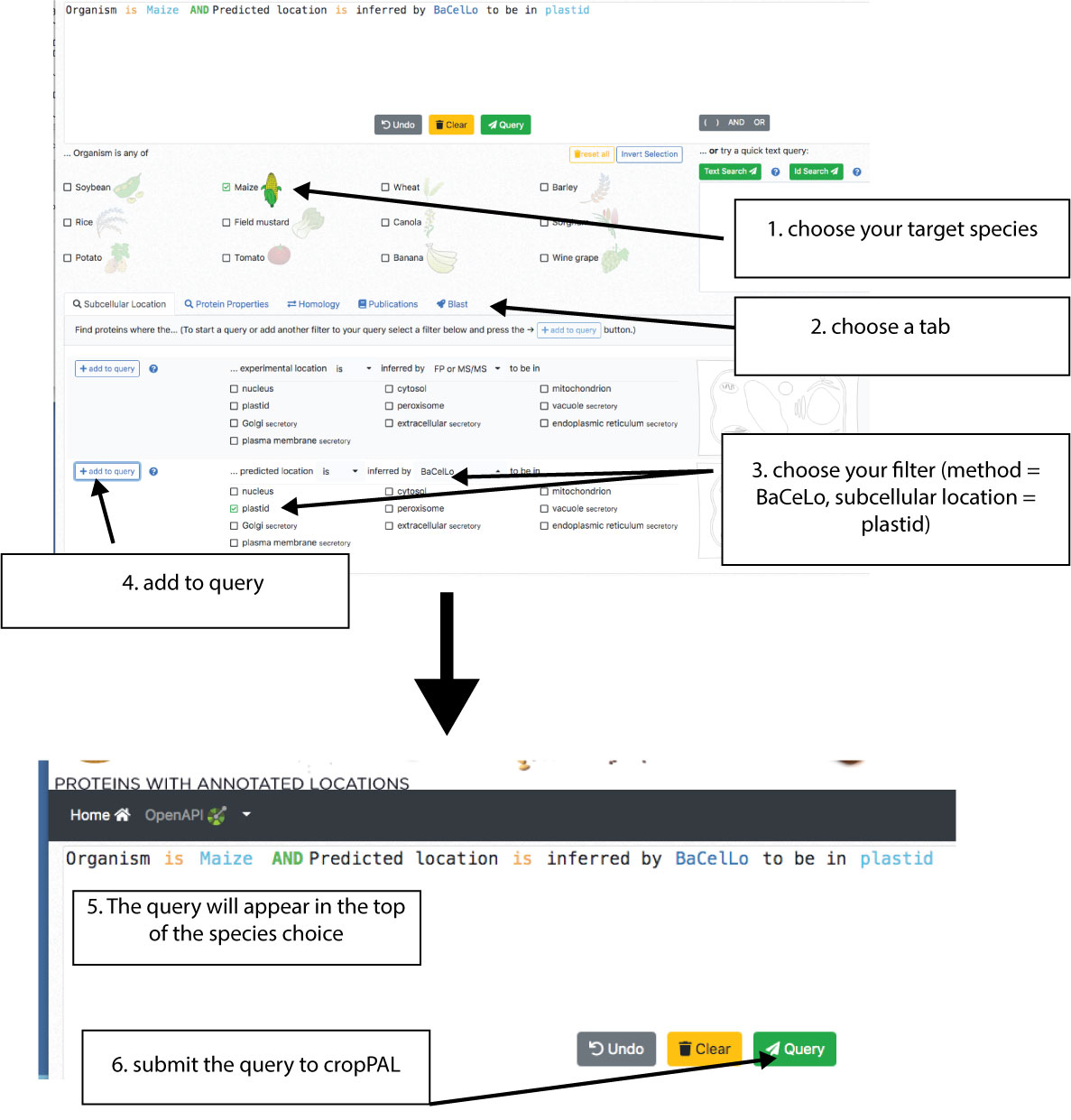
The first step for any search is choosing the crop species that you want to query. The crop species are shown on the left. It is possible to choose one, several or all crops. Users can access the query builder in the lower part of the portal. There are 4 query categories (Subcellular location, Protein Properties, Homology, Publications, BLAST) displayed as tabs. Choose the category of queries (e.g. search for subcellular location) and then choose the filter options (e.g. BaCeLo, plastid). Then add the filter to the query by pressing the add button on the left. This will display query in the top. Use the builder buttons (), AND, OR to add more filters. Once done with the building. Press the “Query” button and retrieve the results.
Search Category
TBD
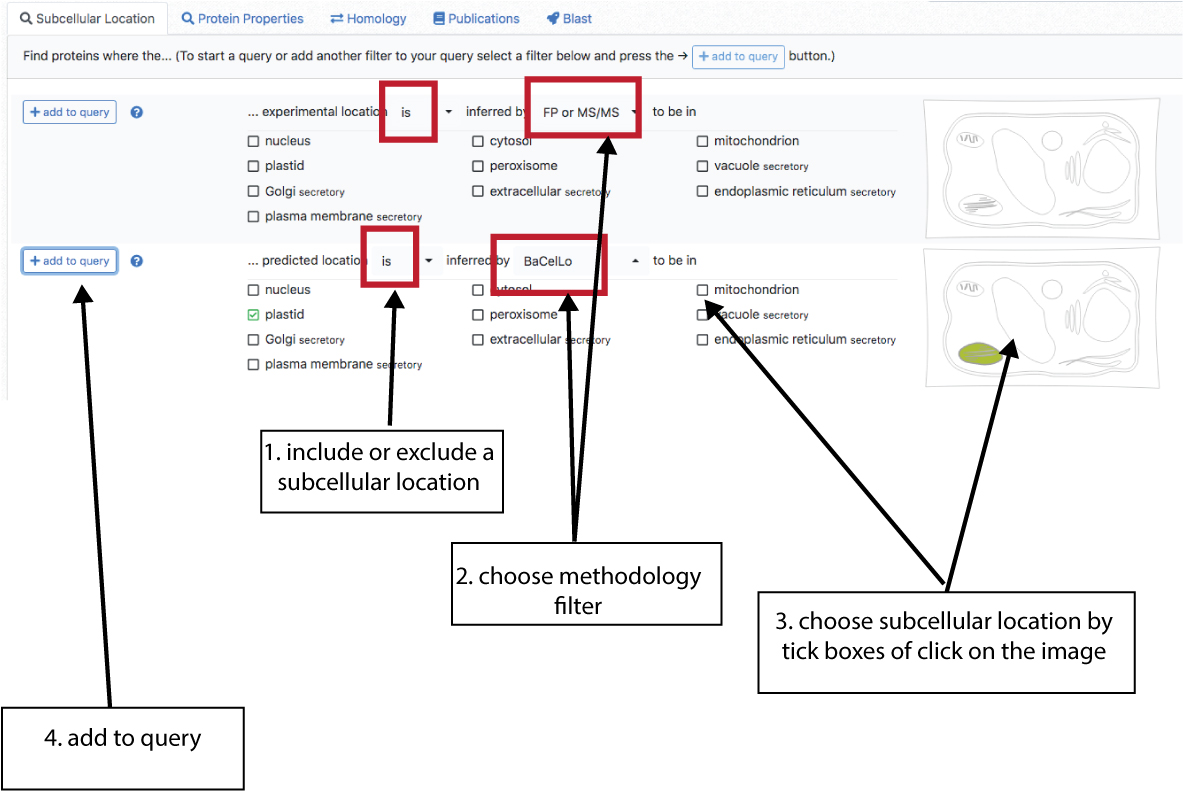
Location Query
This contains filters for subcellular locations. There are two sections, experimental data (top) and predicted data (bottom). The data can be filtered by type of experimental (FP or MSMS or any of the two), predictor (individual predictors, any of them or WTA = a consensus vote) and subcellular location (plastid and,or nucleus and, or …). The drop down bars allow inclusion (“is in location xyz”) or exclusion (“is not in location xyz”) of locations. Subcellular locations can be added using the tick box lists as well as clicking on a location in the cell schematic.
Protein Property Queries
The cropPAL data sets can be filtered for protein properties. This includes protein descriptions (Ensembl annotations), physical protein properties (amino acid length, molecular weight, isoelectric point, GRAVY), chromosome or assembly location of gene encoding the protein, protein features (GO terms, motifs, alias, other IDs…). Choose the filters from a line and press the add button on the left. To choose more than one filter, add one to the query and then press AND/OR in the query builder window at the top. Handy note: The protein feature window on the bottom accepts copy past text of lists of GO terms or other features.
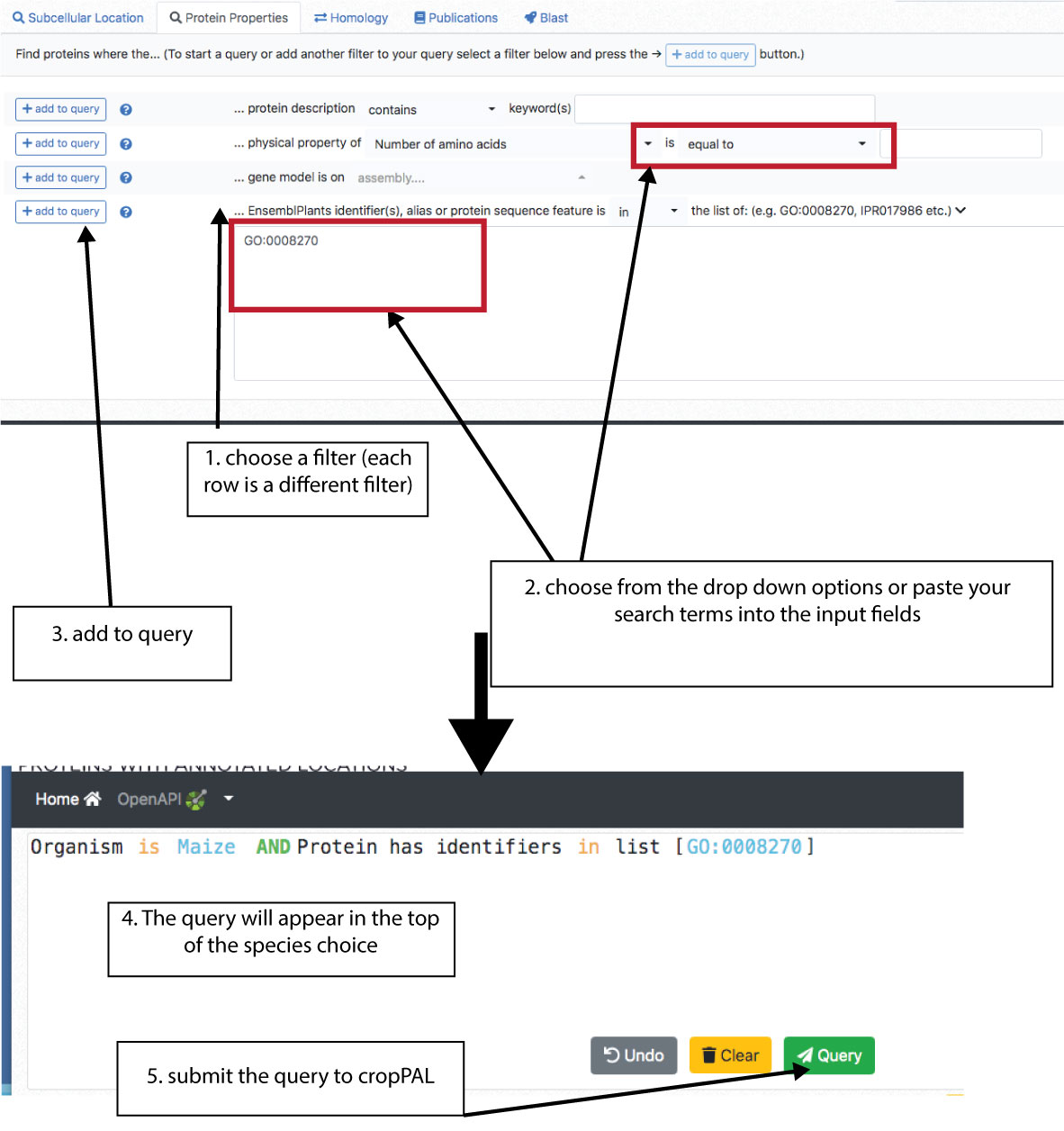
Homology Queries
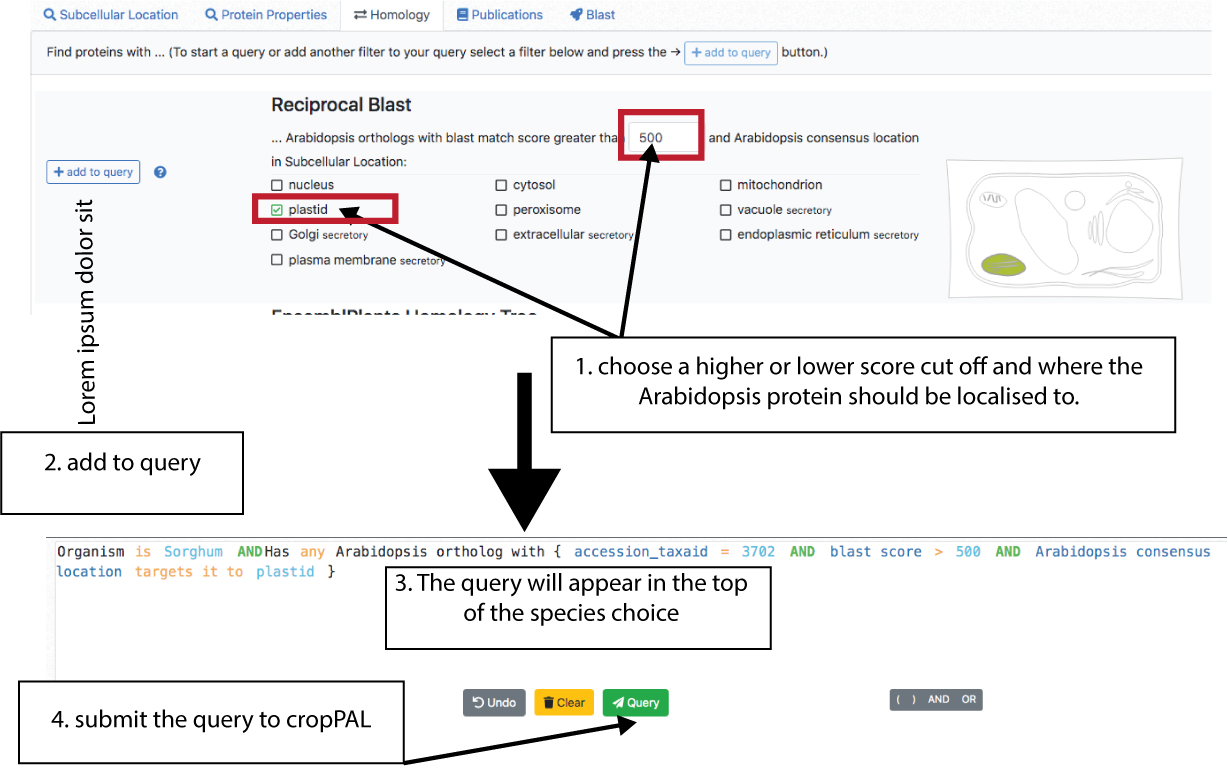
The orthology filters included in cropPAL derive from either reciprocal blasting of crop proteins against Arabidopsis thaliana proteins (TAIR10, top filter) or using the crop species homology tree generated by Ensemble using TreeBEST.
The Reciprocal BLAST option will return all the crop orthologues of Arabidopsis proteins filtered for a chosen subcellular location. For examples trying to find all orthologues in sorghum where the Arabidopsis protein has been classified as a plastid protein (by SUBAcon, a consensus classifier that takes into account experimental and predictions from 22 predictors). The user can set a cut off for similarity for the reciprocal blast score according to personal preferences.
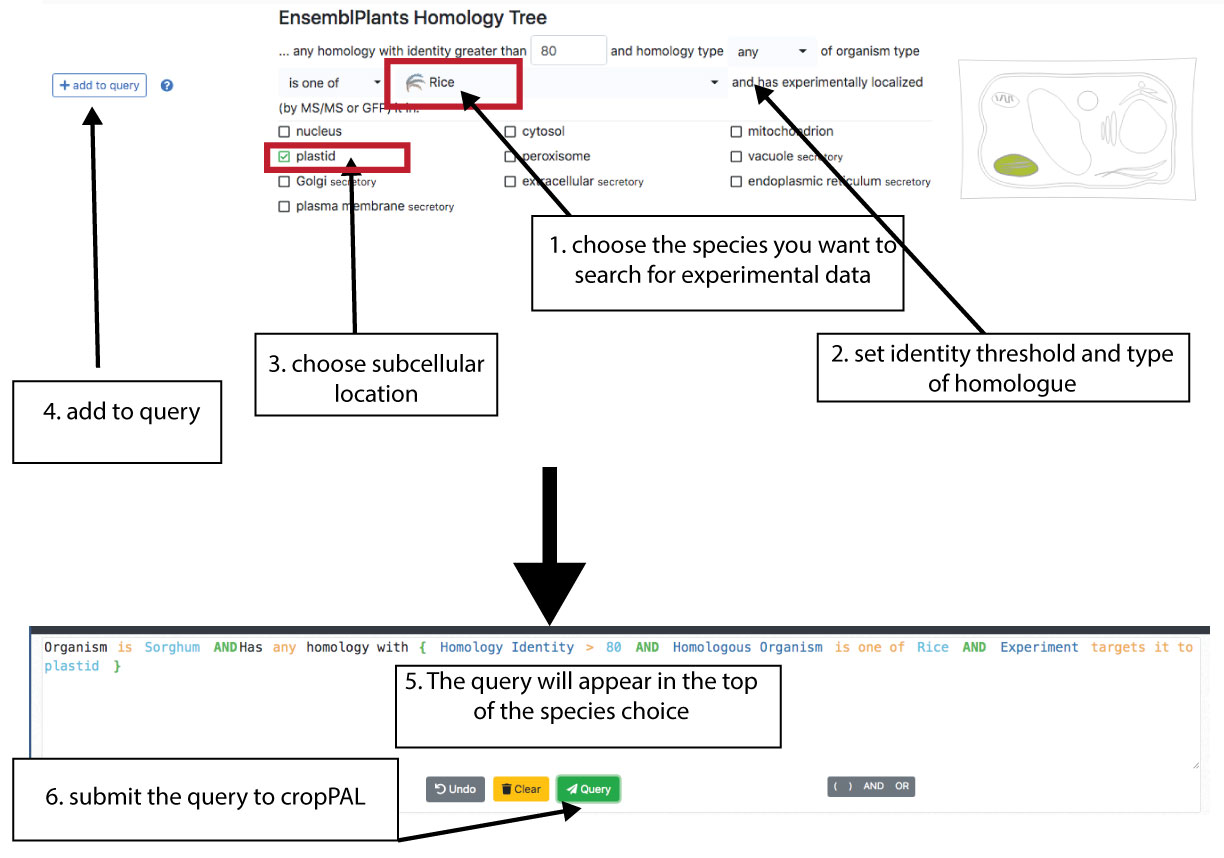
The EnsemblPLants Homology Tree is a Multi-species tree can be used to find orthologues and paralogues with sequence identify where experimental data exists for a another species. This function allows to search any species against any (single or multiple). The homology-tree is more stringent than the reciprocal BLAST. From the example above we searched sorghum against Arabidopsis. With the homology tree we can find all sorghum proteins were homologues proteins in rice have experimental evidence to be located in the plastid.
Note: the homology connections can take a while due to the large amount of links the query has to go through. In particular if several subcellular locations and species are being searched. If you have a very large query and it will not work through our portal please contact us for assistance.
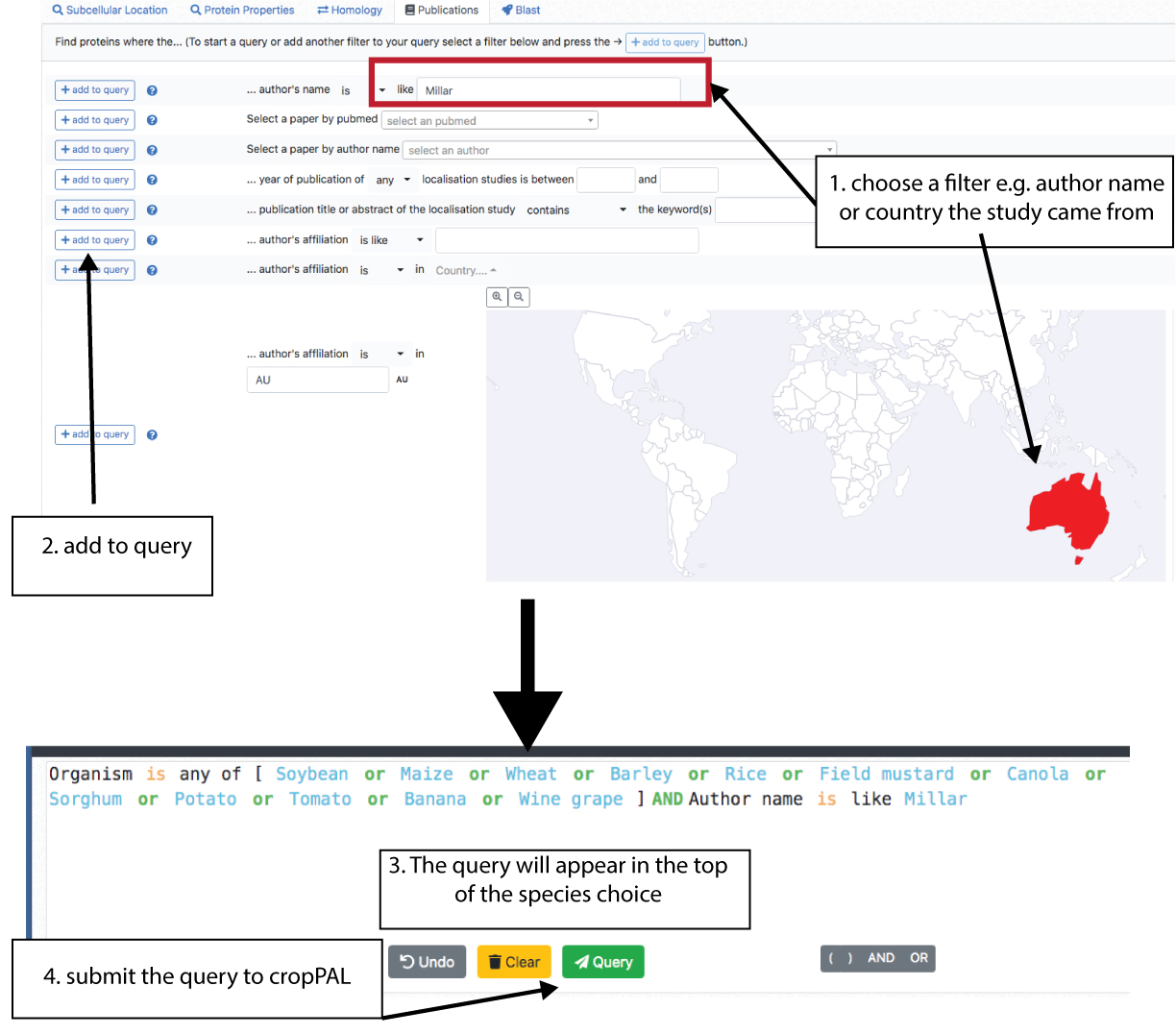
Publications Queries
The Publication filter allows the user to filter cropPAL data for individual experimental studies, the study origin (country, institutions), study author names, year of publication as well as keywords within titles and abstracts of experimental studies.
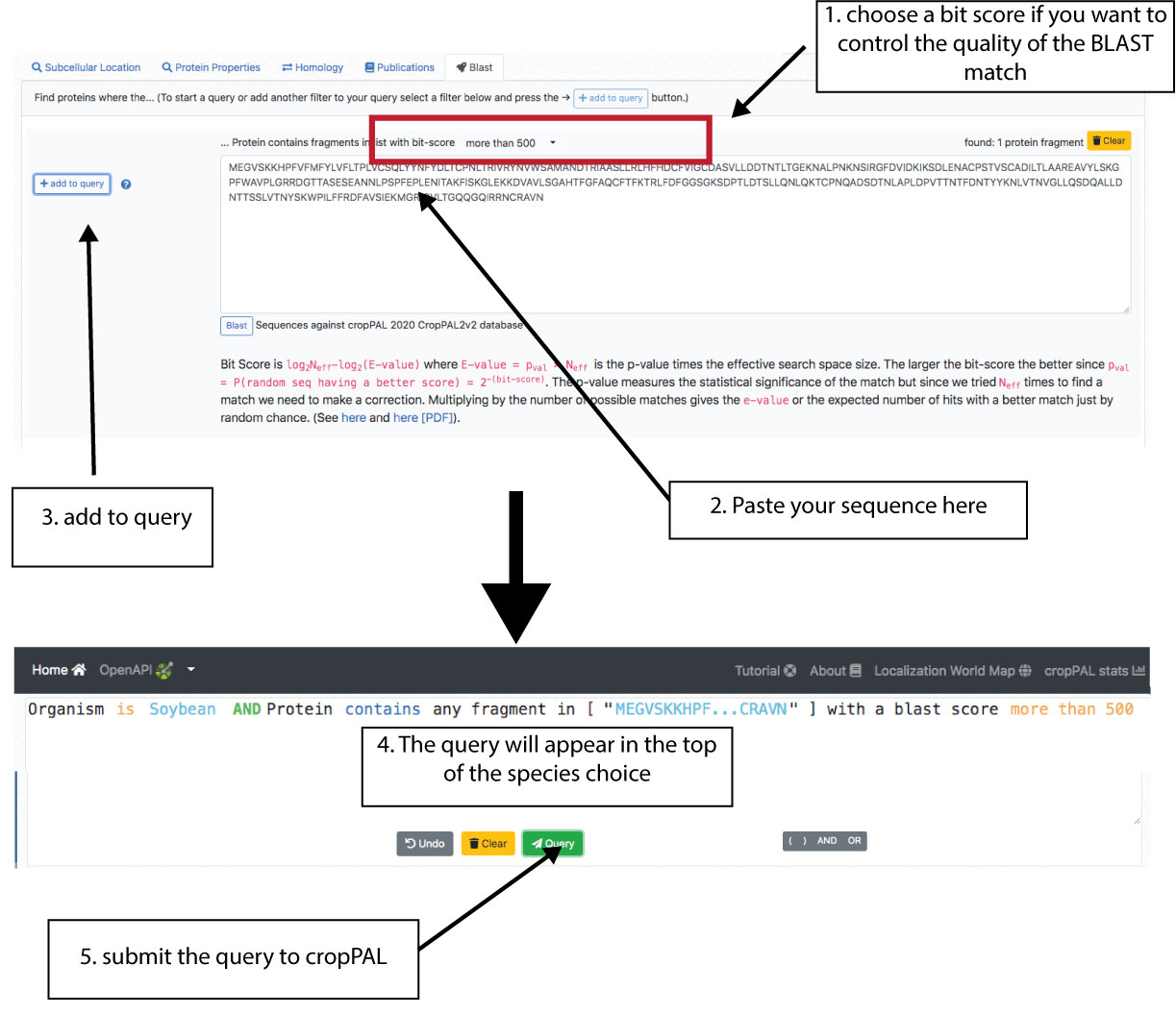
BLAST query
The BLAST tab allows the user to search a sequence against the cropPAL proteomes. Thereafter a user may have a protein sequence of a cropPAL species and cannot link it to an ID that cropPAL can recover. The user can paste the sequence (without breaks and gaps) into the BLAST window and BLAST it against the target species. Alternatively the user can BLAST a sequence from another species not in cropPAL (e.g. chickpea protein). The BLAST will find the closest match in the cropPAL species (e.g. user may chose soybean or all species in cropPAL)
Query stacking
Building multi-filter queries can be achieved by pressing the AND, OR functions underneath the query window in between adding each filter from the Query tabs. The full query will appear in the query window. (See example below using 3 location filter).